IBM today launched ‘COVID notebooks,’ an open-source data solution designed to help overwhelmed developers, researchers, and policy-makers.
When the big-wigs put their heads together to work on pandemic solutions, they need solid datasets full of up-to-date, organized, clean data. But coming up with these datasets is a monumental endeavor.
According to IBM:
The information landscape is overwhelming. A near-constant flow of data from research studies, news outlets, social media, and health organizations make the task of analyzing data into useful action nearly impossible. Developers and data scientists need answers to their questions about data sources, tools, and how to draw meaningful and statistically valid conclusions from the ever-changing data.
Policy makers face similar challenges. The United States has over 3,000 counties, each with a unique story of how COVID-19 is impacting its community.
IBM’s new COVID resource is an open-source Jupyter notebook containing pandemic-related datasets and tools derived from authoritative data sources such as John Hopkins University, The New York Times, and the European Centre for Disease Prevention and Control.
As pandemic data changes on a daily basis, the COVID notebook uses Elyra Notebook Pipelines Visual Editor and KubeFlow Pipelines to ensure researchers have clean, up-to-date datasets.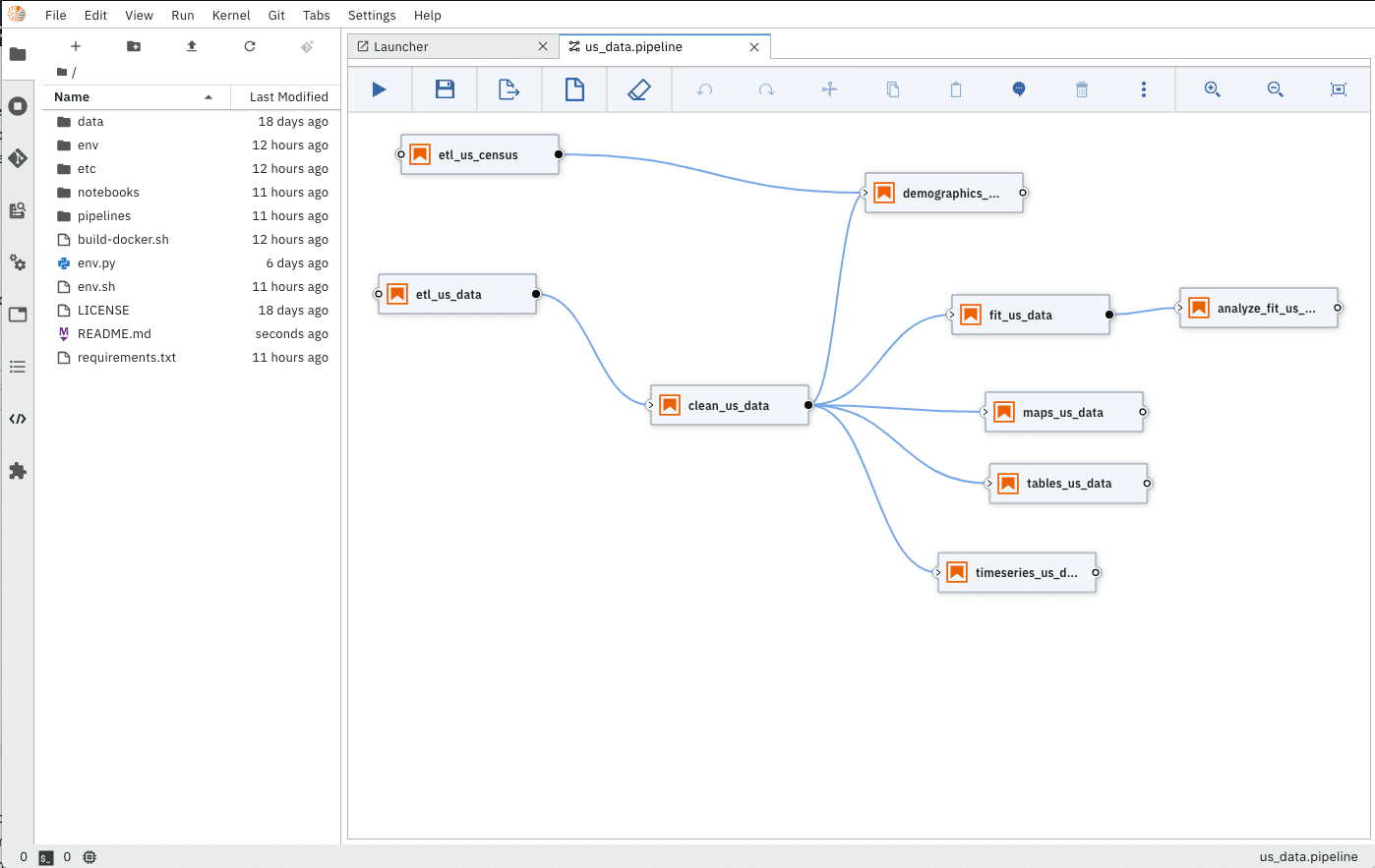
In other words, IBM’s taken as many pain-points out of the data aggregation and implementation process as possible.
This frees developers and researchers up to focus on the tasks of deep analysis and prediction modeling and gives policy-makers quick, easy access to granular geographic details.
For more information check out IBM’s blog post here. You can access COVID notebooks here on GitHub.
Get the TNW newsletter
Get the most important tech news in your inbox each week.